Research
Research intro in motion pictures
Physiological and biophysical principles of cell surfaces
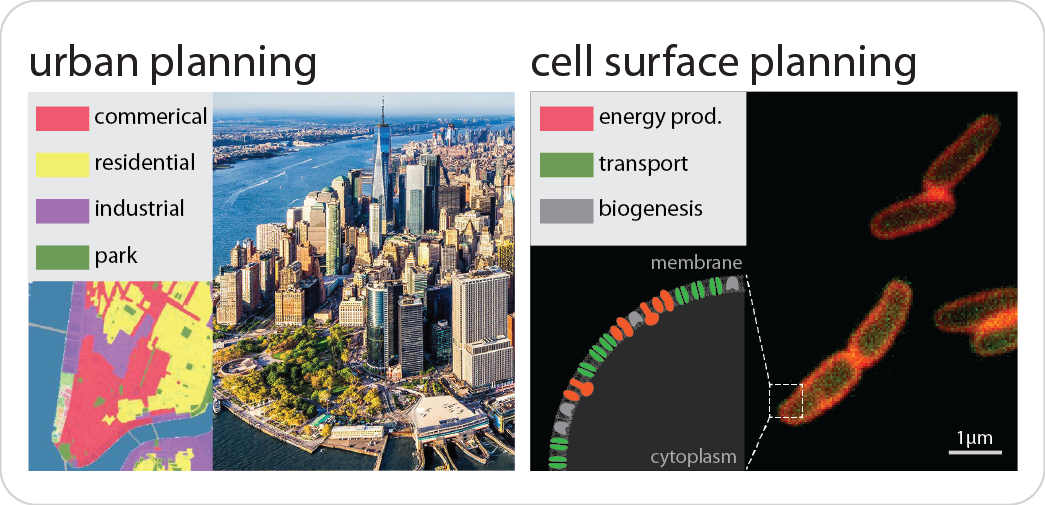
The cell membrane can be seen as a microscopic version of Manhattan, NYC. The cell almost packs the limited membrane space with proteins that provide essential functions. With such high packing density, the cell must evolve to do really good job in “floorplanning” its membrane to optimize the fitness. The quantity and spatial organization of membrane proteins are crucial in determining growth rate, death rate, as well as adaptability in new environments. We are quantifying basic membrane properties using single-cell microfluidics and imaging approaches and performing perturbation experiments to test quantitative models.
Quantitative principles of cellular adaptation
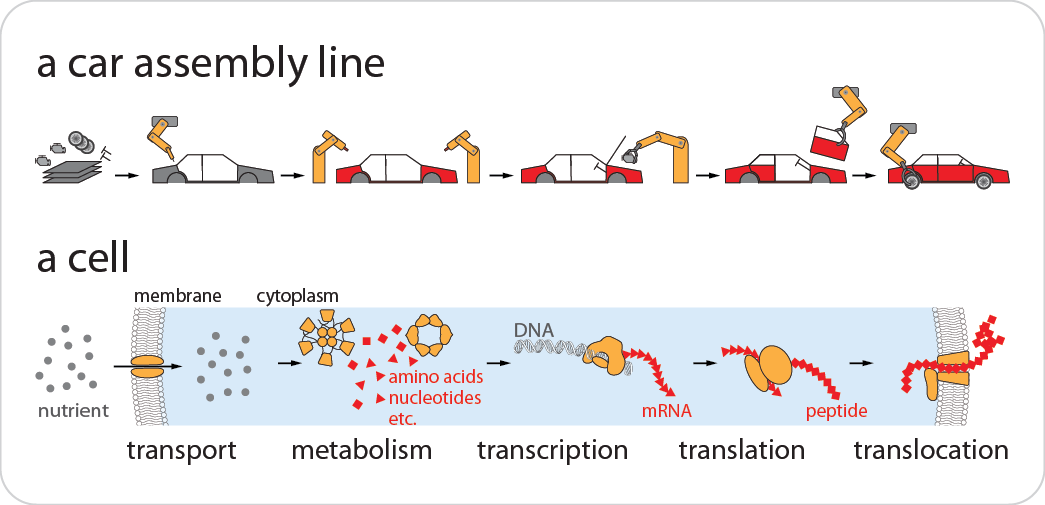
The abundance of many cellular components is often more than a cell needs. For example, a bacterial cell always has extra copies of essential proteins, e.g., ribosomes, or extra physical space, e.g., membrane area – removing some of them does not affect corresponding cellular functions. Why does the cell use such a seemingly wasteful strategy? It is thought that extra components are essential for the cell to adapt to a new environment, and cellular adaptation remains an unresolved fundamental biological problem. We will experimentally gauge how the extra abundance of cellular components is tuned in different environments, hopefully to gain insights into the biological mechanisms underlying cellular adaptation.
Physiological and biophysical principles of bacteria-phage interactions
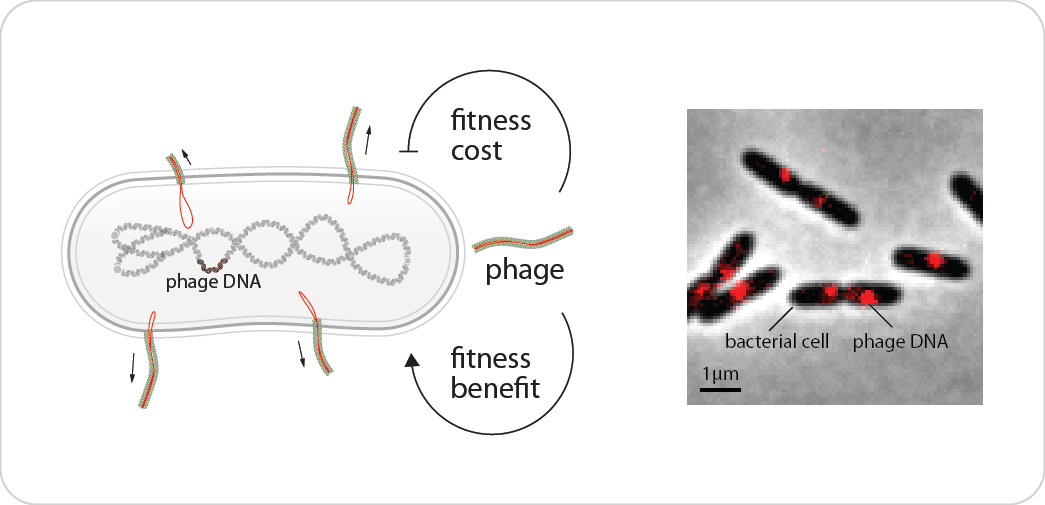
While bacteriophages (viruses that infect bacteria) are often known for killing their host, a large class of bacteriophage are very temperate – they live quite peacefully with their bacterial host, and do not kill when replicating and leaving the host. It is even reported that these phages benefit their host to survive environmental challenges at least at the community level. However, the fitness benefit and cost of phage production in bacterial host is poorly understood. We are trying to delineate the physiological consequences (e.g. growth, survival and adaptability to new environments) of the bacterial-phage interactions. This is also a great opportunity to bridge the quantitative principles of microbial growth at both single-cell and community levels.
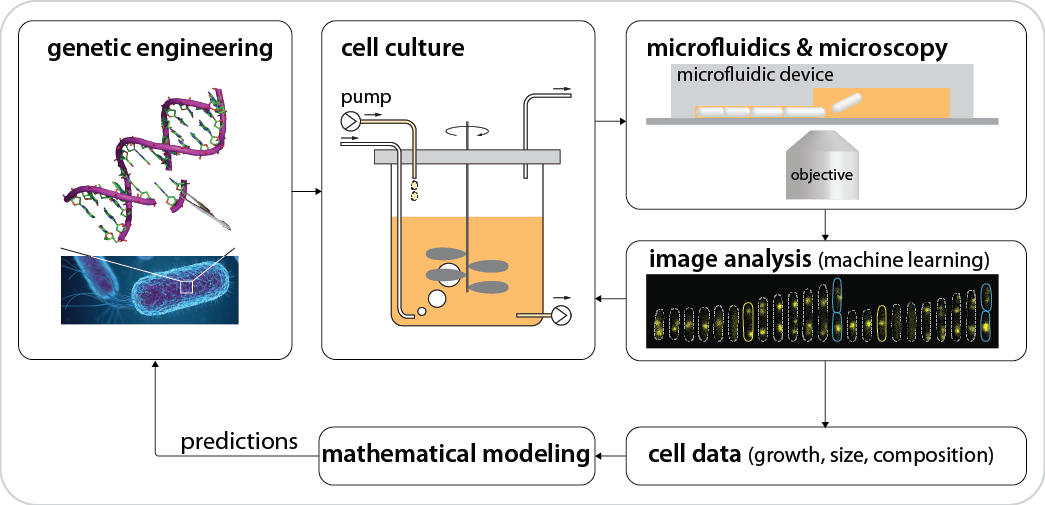
Tools used (skills you can learn) in our lab
We develop and adapt multidisciplinary tools, such as genetic engineering, cell culture, microfluidics, microscopy, image and data analysis, machine learning, and physical modeling. These tools form a closed-loop pipeline to allow us to iterate between quantitative predictions and measurements by refining models and redesigning experiments, respectively.